Microbiomes of insect vectors
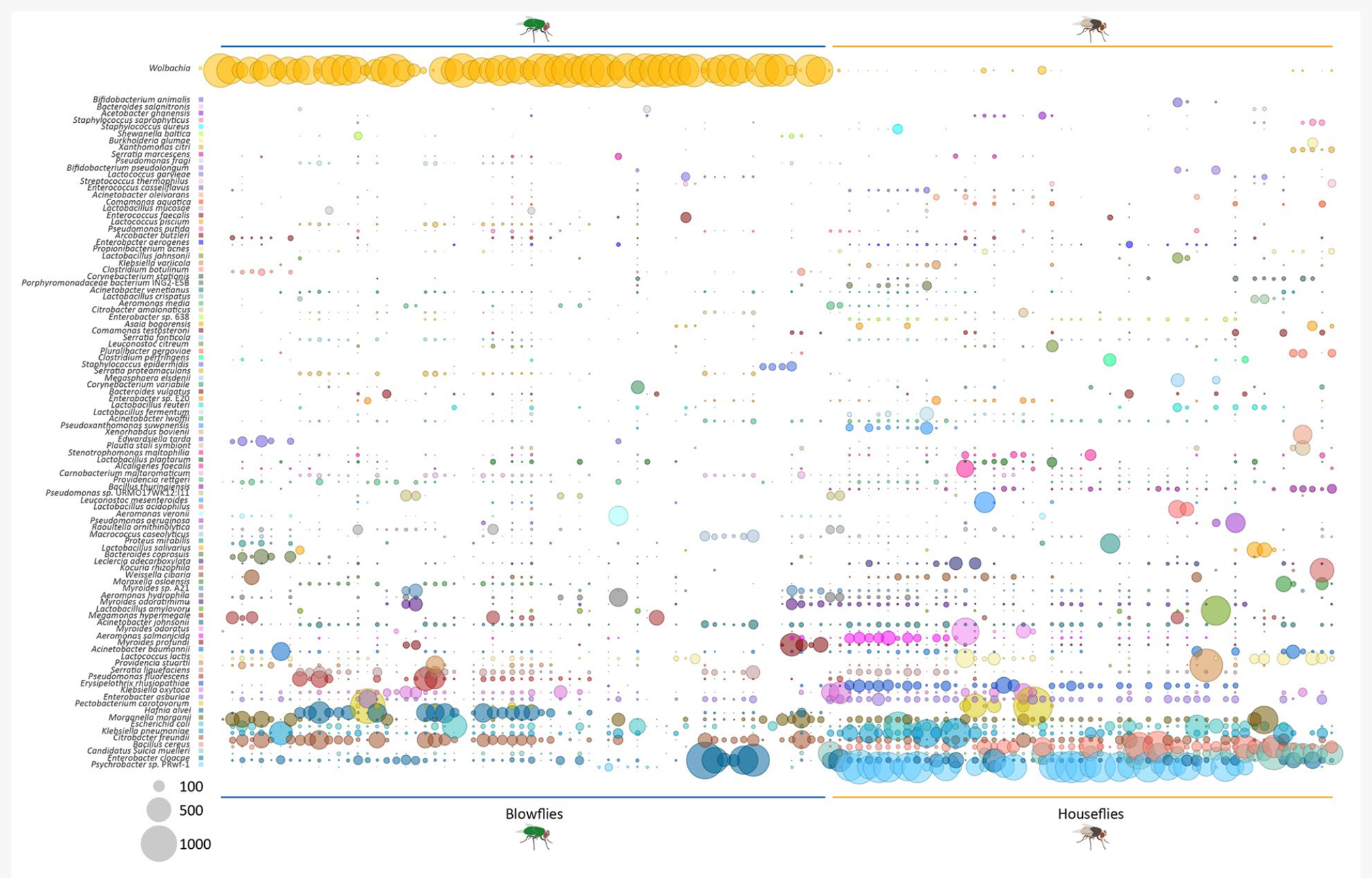
Interactions between hosts and microorganisms are ubiquitous in nature, capable of modulating animal physiology, fitness, protection against infections, and host behavior. We are using whole shotgun metagenomics to study the microbial communities associated with biological (mosquitoes and kissing bugs) and mechanical insect vectors (houseflies and blowflies). Insects are major agents of infectious diseases, often inhabiting densely populated areas where their propagation is increased. Switches in microbial communities could lead us to a set of naturally occurring microbes that respond to metabolic pathways that could help us to assemble a microbiome-based strategy to control vectors of diseases in urban and rural areas. Junqueira et al., 2017. https://www.nature.com/articles/s41598-017-16353-x
Genomics of flies
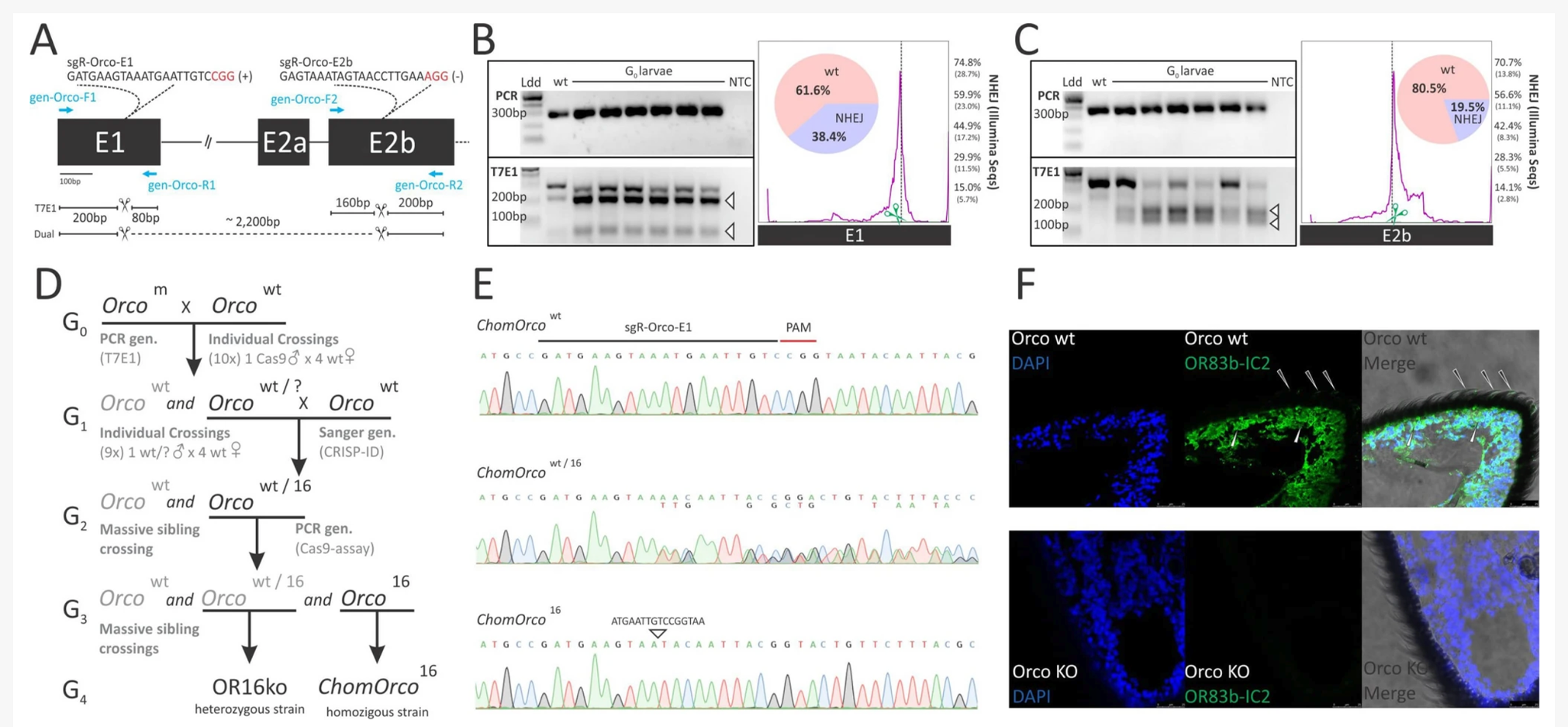
Recent developments in comparative genomics allow us to analyze features that help us understand how genomes evolved, giving rise to various insect habits. In flies (Diptera), different feeding strategies are also associated with the evolution of parasitism and adaptation to living hosts. We are currently working with two genomes of flies generated: the blowfly Chrysomya megacephala and the New World screwworm (NWS) fly Cochliomyia hominivorax. While the blowfly has a necrophagous habit and acts as a vector of diseases, the NWS fly feeds on the living tissues of vertebrate hosts, being one of the most important livestock pests in the New World. Genomic resources generated are also helpful in assisting future developments in vector and pest control based on targeted genome editing using CRISPR/Cas9. Paulo et al., 2021. https://www.nature.com/articles/s41598-021-90649-x
Phylogenomics and Evolution
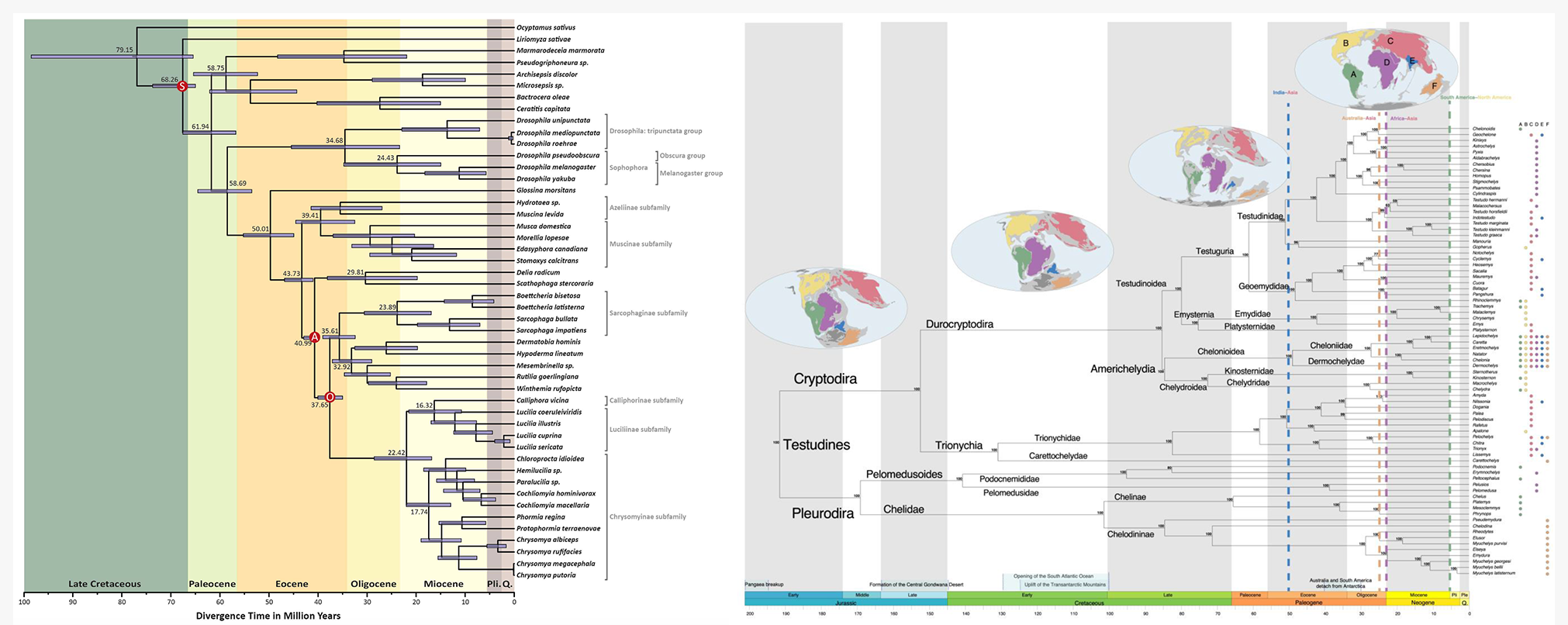
With the onset of new sequencing platforms, an increasing number of genomes became available in public databases and can now be analyzed with methods that deal with the intersection of phylogenetics and genomics. Our research conciliates molecular data with fossil records, geological events, and cutting-edge bioinformatics to estimate the time tree of species in different groups, such as flies and turtles. Junqueira et al., 2016. https://www.nature.com/articles/srep21762. Selvatti et al., 2023. https://doi.org/10.1016/j.ympev.2023.107773.
Environmental metagenomics
The World Health Organization has listed air pollution, climate change, and the emergence of antimicrobial resistance as the main threats to global health in the future. We use breakthrough methods in ultra-low biomass sequencing to analyze the urban microbiome from surfaces swabbed in Rio de Janeiro to uncover the microorganisms circulating in populated areas and their public health impact on future outbreaks. Likewise, we are involved in an integrated analysis of air microbiomes sampled in different locations on the planet, together with extensive meteorological and physicochemical records, which will give insights into the airborne ecosystem. Gusareva et a., 2019. https://www.pnas.org/doi/10.1073/pnas.1908493116